
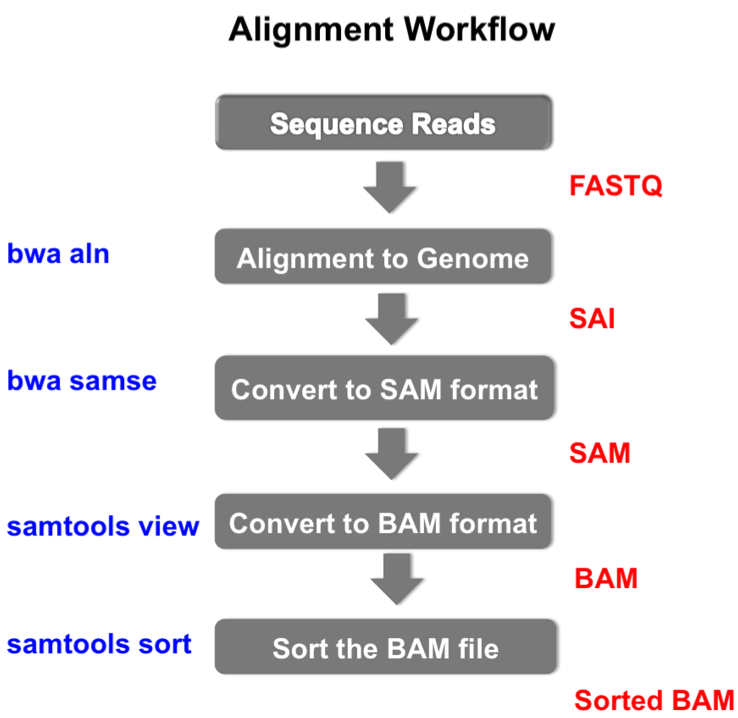
There are 38917 one-transcript genes in the gtf file There are 199184 distinct transcript ID in the gtf file There are 60675 distinct gene ID in the gtf file :~/Documents/rMATS.4.0.2$ python rMATS-turbo-Linux-UCS4/rmats.py -b1 ~/Documents/rMATS.4.0.2/testData/b1.txt -b2 ~/Documents/rMATS.4.0.2/testData/b2.txt -gtf ~/Documents/rMATS.4.0.2/Homo_sapiens.GRCh38.83.gtf -od bam_test -t paired -readLength 50 -cstat 0.0001 -libType fr-unstranded There are 4065 alt 3 SS events and 3087 alt 5 SS events.

There are 3772 one-transcript genes with only one exon in the transcriptĪverage number of transcripts per gene is 2.199698Īverage number of exons per transcript is 9.189876Īverage number of exons per transcript excluding one-exon tx is 10.083076Īverage number of gene per geneGroup is 261.649536įail to open /cluster/tufts/dmcb_kuperwasser02/Jess/STAR_output/4181_mix-NTC19_S1_įail to open /cluster/tufts/dmcb_kuperwasser02/Jess/STAR_output/4185_mix-NTC19_S5_įail to open /cluster/tufts/dmcb_kuperwasser02/Jess/STAR_output/4182_mix-FUBP1Polyclonal_S2_įail to open /cluster/tufts/dmcb_kuperwasser02/Jess/STAR_output/4186_mix-FUBP1Polyclonal_S6_ĭone processing each gene from dictionary to compile AS events There are 5729 one-exon transcripts in the gtf file There are 15188 one-transcript genes in the gtf file
There are 58259 distinct transcript ID in the gtf file There are 26485 distinct gene ID in the gtf file I am running rMATS.4.0.1 on my university's cluster and it look like the program is running until I get the following message:


 0 kommentar(er)
0 kommentar(er)
